Research Areas
At the Devreotes Lab, we study how cells polarize and migrate, either spontaneously or under external cues. We identify different genetic and biochemical components of signal transduction and cytoskeletal networks and dissect how they coordinate to define the "logic" of the circuitry, which in turn, regulates chemotaxis, random migration, macropinocytosis, and other associated cellular processes. In the past, our studies elucidated the intercellular communication process that Dictyostelium uses to self-organize in response to external cues. We identified the first family of chemoattractant receptors that were demonstrated to mediate chemotaxis and cell-cell signaling. We discovered that phosphoinositides play a key role in chemotaxis and in defining the basic strategy that eukaryotic cells use to sense gradients and spatiotemporally organize signaling and cytoskeletal events. We were the first to demonstrate that shallow spatial gradients in G-protein activation leads to dramatic amplification of PI3K activity at the leading edge of the migrating cells. We introduced multiple new conceptual advances to cell migration and polarity field, including LEGI or Local Excitation Global Inhibition model of directional sensing and adaptation, biased coupled excitable network model of autonomous signal transduction and chemotaxis, and biophysical models of plasma membrane self-organization that dynamically regulate polarity in migrating cells.
We use Dictyostelium discoideum amoeba, mammalian leukocytes (macrophages and neutrophils), and different cancer cell lines as our model systems. We extensively use cutting-edge cellular engineering approaches, synthetic biological and optogenetic tools, and high-end microscopy techniques, in conjunction with stochastic computational models. In the process, we also developed multiple new tools to perturb specific genetic/biochemical pathways (and even nonspecific biophysical properties), in a spatiotemporally controlled fashion. We are eager to learn all the fundamental aspects of cell migration and polarity. A comprehensive understanding of these fascinating cellular processes would be the key to developing therapies for diverse pathologies such as autoimmune diseases, cardiovascular diseases, cognitive deficits, and cancer metastasis. Please visit our research page to learn more. The recent areas of interest are provided below.
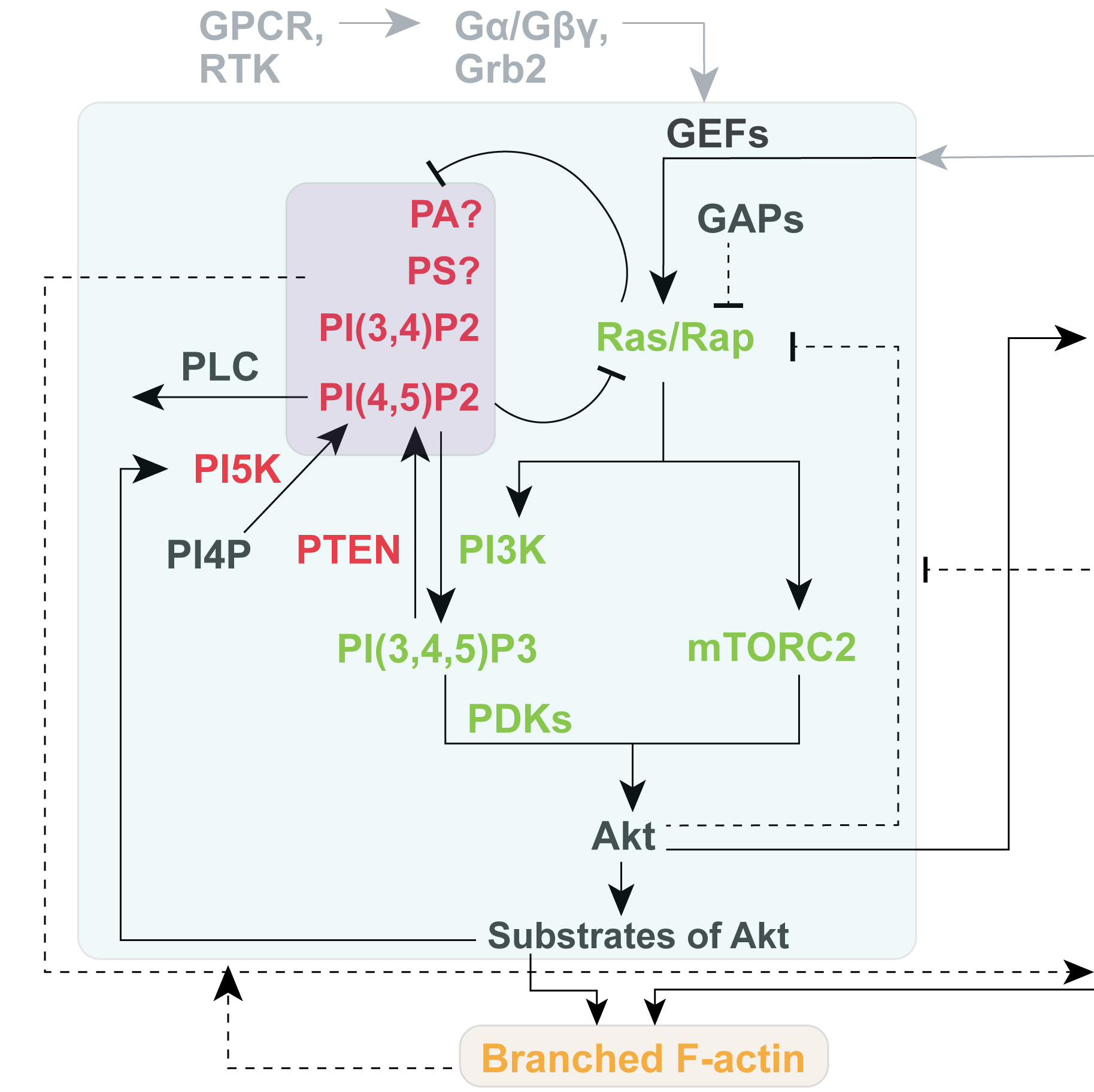
Genetic and Systemic Dissection of Signaling Network
Which genetic components are involved in specific signal transduction cascades? How do cells coordinate numerous signaling and cytoskeletal events via internal feedback loops? How do cells decide when and where to activate a specific signaling event so that they can polarize and migrate correctly? How do cells resolve different conflicting environmental cues and determine how to spatiotemporally regulate stochastic activities? These are some questions we are focused on.
Learn More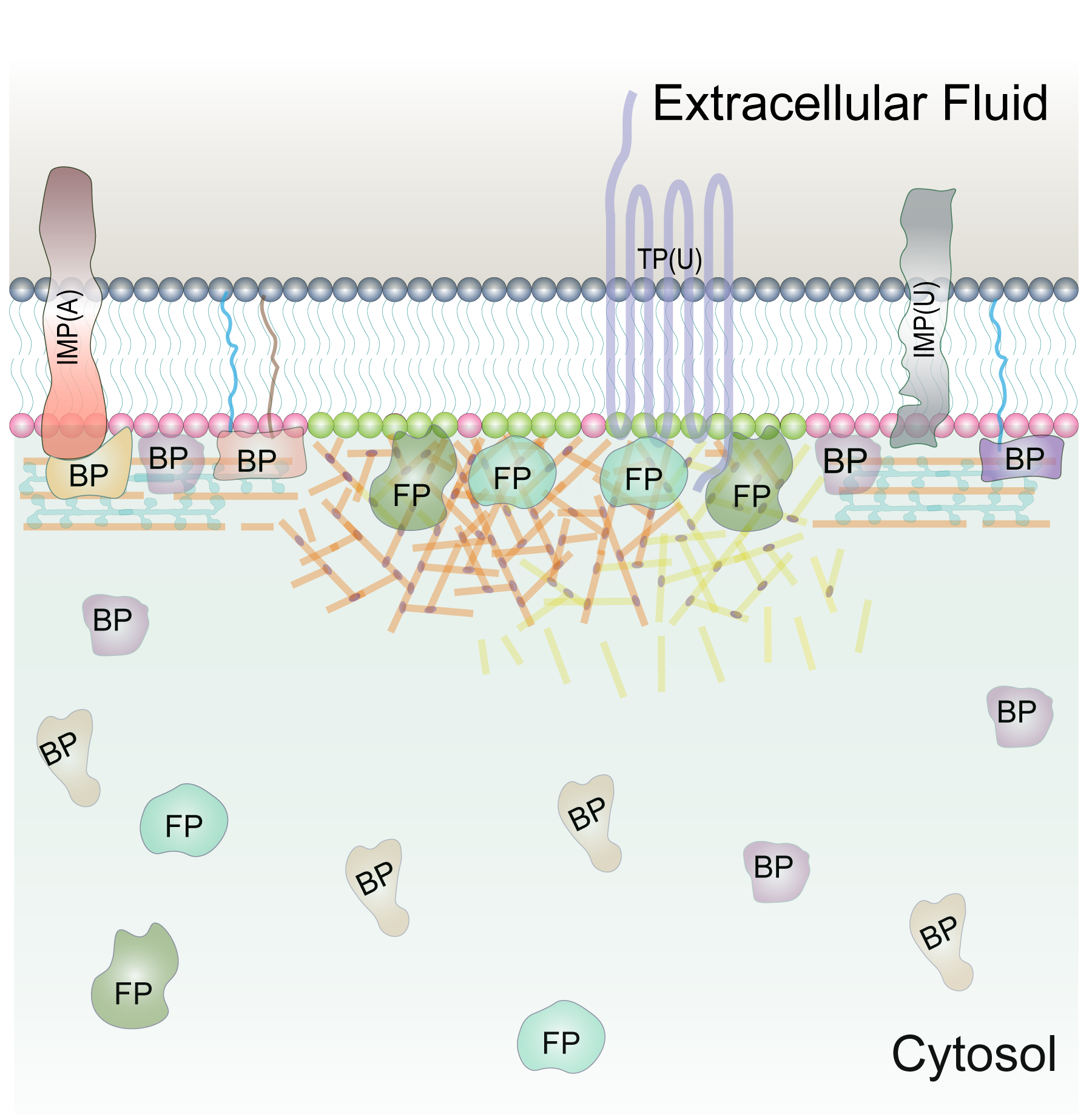
Membrane Biophysics and Lipid Biology of Migrating Cell
How does the plasma membrane acts as the hub of signal transduction during cell migration and polarity? How do different classes of membrane proteins dynamically compartmentalize into specific membrane regions? How is the symmetry breaking process initiated in plasma membrane? How do different lipid compositions and biophysical properties of membrane domains define the signaling state? How does the plasma membrane integrate information to encode templates for self-organization? Our research aims to address these questions.
Learn More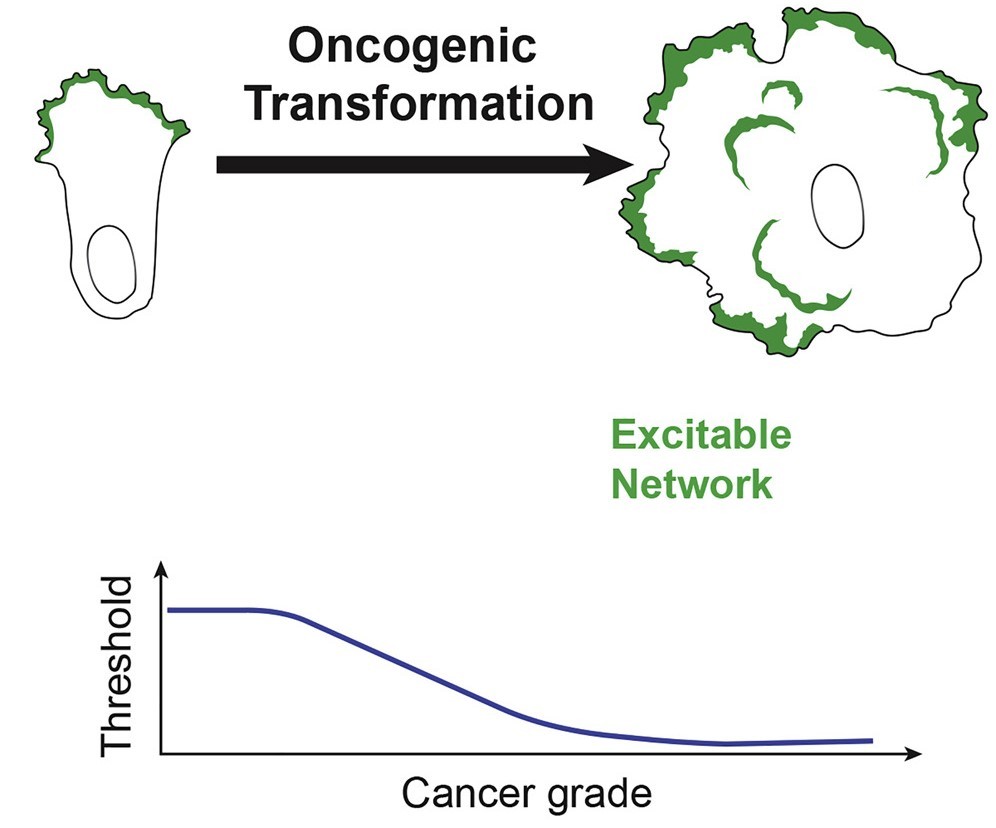
Cell Biology of Oncogenesis and Cancer metastasis
Which alterations in polarity signaling pathways result in oncogenesis? How do defects in cell migration transform normal cells to cancer cells? How do cancer cells differ from normal cells in terms of the spatiotemporal organization of signaling and cytoskeletal events? Do these organizational changes reprogram the energetics and metabolism of cancer cells? We are really interested in finding answers to these questions.
Learn MoreMeet the Team
We have maintained a long-term collaboration with computational biologist Pablo A Iglesias and his laboratory.
While backgrounds vary widely covering a large spectrum of life sciences and engineering, lab members lab members share a passion to learn n ew concepts as well as apply their expertise to solve fundamental problems in cell migration that can eventually help in developing advanced therapeutics.
To view the profiles of the current graduate students, postdoctoral fellows, and research staffs, please click here. The list of alumni is also available.
If you think you would be a good fit and would like to join the team, please feel free to contact us.

News from the Devreotes lab
See the full news achieve here.
Recent Publication Highlights
- Lin Y*, Pal DS*, Banerjee P, Banerjee T, Qin G, Deng Y, Borleis J, Iglesias PA and Devreotes PN. 2024. Ras suppression potentiates rear actomyosin contractility-driven cell polarization and migration. Nat Cell Biol. 26(7):1062-1076. DOI: 10.1038/s41556-024-01453-4.
- Banerjee T, Matsuoka S, Biswas D, Miao Y, Pal DS, Kamimura Y, Ueda M, Devreotes PN, and Iglesias PA. 2023. A dynamic partitioning mechanism polarizes membrane protein distribution. Nat Commun. 14:7909. DOI: 10.1038/s41467-023-43615-2.
- Pal DS, Banerjee T, Lin Y, de Trogoff F, Borleis J, Iglesias PA, Devreotes PN. 2023. Actuation of single downstream nodes in growth factor network steers immune cell migration. Dev Cell 58(13):1170-1188.e7. DOI: 10.1016/j.devcel.2023.04.019
- Yang Q, Miao Y, Banerjee P, Hourwitz MJ, Hu M, Qing Q, Iglesias PA, Fourkas JG, and Losert W, and Devreotes PN. 2022. Nanotopography modulates intracellular excitable systems through cytoskeleton actuation. Proc. Natl. Acad. Sci. U.S.A. 120(19): e2218906120 DOI:10.1101/2022.06.09.495528.
- Banerjee T, Biswas D*, Pal DS*, Miao Y, Iglesias PA, and Devreotes PN. 2022. Spatiotemporal dynamics of membrane surface charge regulates cell polarity and migration. Nat Cell Biol. 24, 1499–1515 (2022). DOI: 10.1101/2022.05.19.492577.
See the entire publication list here.